Preamble
import numpy as np # for multi-dimensional containers
import pandas as pd # for DataFrames
import plotly.graph_objects as go # for data visualisation
import plotly.express as px
Introduction
Before the main optimisation process (the "generational loop") can begin, we need to complete the initialisation stage of the algorithm. Typically, this involves generating the initial population of solutions by randomly sampling the search-space. We can see in the figure below that this initialisation stage is the first real stage, and it's only executed once. There are many schemes for generating the initial population, and some even include simply loading in a population from an earlier run of an algorithm.
Randomly sampling the search-space
When generating an initial population, it's often desirable to have a diverse representation of the search space. This supports better exploitation of problem variables earlier on in the search, without having to rely solely on exploration operators.
We previously defined a solution
We also defined a multi-objective function
However, we need to have a closer look at how we describe a general multi-objective optimisation problem before we initialise our initial population.
We may already be familiar with some parts of Equation 3, but there are some we haven't covered yet. There are
The lower
D_lower = [-2, -2, -2, 0, -5, 0.5, 1, 1, 0, 1]
D_upper = [ 1, 2, 3, 1, .5, 2.5, 5, 5, 8, 2]
In Python, we normally use np.random.rand() to generate random numbers. If we want to generate a population of 20 solutions, each with 10 problem variables (
D = 10
population = pd.DataFrame(np.random.rand(20,D))
population
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.256329 | 0.379697 | 0.551219 | 0.913193 | 0.857961 | 0.230993 | 0.498392 | 0.318684 | 0.954690 | 0.040283 |
| 1 | 0.597399 | 0.111841 | 0.801090 | 0.492588 | 0.211586 | 0.153679 | 0.448325 | 0.696537 | 0.541425 | 0.495963 |
| 2 | 0.461949 | 0.998844 | 0.898229 | 0.937838 | 0.064428 | 0.182616 | 0.076086 | 0.976655 | 0.537354 | 0.736609 |
| 3 | 0.539136 | 0.748668 | 0.570238 | 0.476345 | 0.547363 | 0.527805 | 0.283425 | 0.533672 | 0.126736 | 0.006231 |
| 4 | 0.899781 | 0.910640 | 0.054392 | 0.933451 | 0.318863 | 0.070565 | 0.425399 | 0.615656 | 0.762153 | 0.840374 |
| 5 | 0.034428 | 0.257122 | 0.719380 | 0.602291 | 0.347610 | 0.278748 | 0.480196 | 0.430053 | 0.740159 | 0.241565 |
| 6 | 0.293102 | 0.341901 | 0.020379 | 0.103552 | 0.651027 | 0.824640 | 0.228864 | 0.771375 | 0.260334 | 0.348736 |
| 7 | 0.680644 | 0.586685 | 0.133200 | 0.486397 | 0.350765 | 0.089453 | 0.068280 | 0.777695 | 0.009198 | 0.454770 |
| 8 | 0.711749 | 0.594902 | 0.744637 | 0.228525 | 0.143319 | 0.827422 | 0.578844 | 0.671773 | 0.906182 | 0.044869 |
| 9 | 0.891744 | 0.014426 | 0.737016 | 0.466329 | 0.676474 | 0.623777 | 0.288000 | 0.367927 | 0.590480 | 0.807840 |
| 10 | 0.638020 | 0.671006 | 0.258851 | 0.364851 | 0.896198 | 0.886503 | 0.616957 | 0.153707 | 0.678438 | 0.273749 |
| 11 | 0.243913 | 0.399417 | 0.661920 | 0.676458 | 0.511127 | 0.068651 | 0.627802 | 0.256262 | 0.224177 | 0.397801 |
| 12 | 0.853520 | 0.807107 | 0.989675 | 0.324906 | 0.967510 | 0.029888 | 0.829780 | 0.186981 | 0.945017 | 0.603155 |
| 13 | 0.257897 | 0.777435 | 0.635757 | 0.438057 | 0.701193 | 0.304976 | 0.600306 | 0.684364 | 0.457293 | 0.385380 |
| 14 | 0.015805 | 0.282475 | 0.089203 | 0.645140 | 0.870852 | 0.113519 | 0.550223 | 0.548623 | 0.683554 | 0.970726 |
| 15 | 0.750482 | 0.351151 | 0.163974 | 0.395036 | 0.487530 | 0.828154 | 0.400699 | 0.874083 | 0.127836 | 0.930238 |
| 16 | 0.275726 | 0.613227 | 0.143559 | 0.281961 | 0.931625 | 0.038933 | 0.402476 | 0.388847 | 0.927828 | 0.847508 |
| 17 | 0.912328 | 0.885590 | 0.338265 | 0.893919 | 0.848467 | 0.837799 | 0.007764 | 0.500073 | 0.403926 | 0.109104 |
| 18 | 0.938348 | 0.628789 | 0.062532 | 0.836181 | 0.600204 | 0.939957 | 0.546433 | 0.169062 | 0.607440 | 0.543939 |
| 19 | 0.778387 | 0.730593 | 0.626103 | 0.293385 | 0.417516 | 0.675058 | 0.449187 | 0.772960 | 0.563766 | 0.720014 |
This works fine if all of our problem variables are to be within the boundaries 0 and 1 (np.random.uniform() instead.
population = pd.DataFrame(
np.random.uniform(low=D_lower, high=D_upper, size=(20, D))
)
population
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.937522 | -0.467837 | 1.152330 | 0.846117 | -3.959224 | 1.111467 | 1.764888 | 2.023382 | 5.017223 | 1.754590 |
| 1 | 0.057486 | -0.107231 | -1.535716 | 0.860808 | 0.092036 | 0.895447 | 1.594686 | 1.765635 | 6.273598 | 1.499652 |
| 2 | -0.080653 | -1.739054 | 1.723938 | 0.278877 | -1.580010 | 0.655388 | 4.748730 | 4.651188 | 6.928463 | 1.539721 |
| 3 | -1.373899 | -0.758680 | -0.955178 | 0.093807 | -3.732337 | 1.524556 | 4.539603 | 1.810522 | 7.959026 | 1.800988 |
| 4 | -0.717957 | -0.251662 | 1.566245 | 0.549718 | -4.317165 | 2.354138 | 2.966314 | 2.558895 | 0.147347 | 1.303226 |
| 5 | -0.064253 | -1.837954 | 0.648241 | 0.813850 | 0.425318 | 1.586562 | 3.213497 | 4.088906 | 4.735539 | 1.934544 |
| 6 | -0.011683 | -1.557681 | -0.998977 | 0.796972 | -0.715962 | 0.758970 | 1.870444 | 4.598365 | 7.665607 | 1.313399 |
| 7 | -0.553515 | -0.805990 | 2.112900 | 0.164496 | -0.602322 | 1.367240 | 1.964215 | 2.634935 | 1.089449 | 1.470806 |
| 8 | -0.869776 | -1.162846 | -1.144249 | 0.794074 | -0.529364 | 1.496111 | 3.582352 | 4.778561 | 1.860098 | 1.605092 |
| 9 | -1.770820 | 0.285399 | 2.172822 | 0.261950 | 0.446716 | 1.498100 | 2.007516 | 4.571324 | 7.319199 | 1.000806 |
| 10 | 0.624058 | -1.782481 | 0.467486 | 0.128657 | -3.014476 | 1.435527 | 3.107921 | 4.939729 | 3.500721 | 1.780649 |
| 11 | 0.237057 | 1.446570 | 2.342194 | 0.269955 | -2.960841 | 1.848730 | 1.258174 | 4.097886 | 7.271046 | 1.291888 |
| 12 | -1.449719 | -1.135843 | -0.867221 | 0.328213 | -2.342167 | 0.906001 | 4.518713 | 4.281935 | 0.868588 | 1.387121 |
| 13 | 0.535788 | -1.252023 | -1.403047 | 0.919144 | -0.766802 | 0.860278 | 2.432873 | 2.292833 | 3.336979 | 1.573485 |
| 14 | -1.876729 | -1.556136 | 0.481709 | 0.133905 | -3.106224 | 0.733688 | 3.448770 | 1.564956 | 0.618818 | 1.694889 |
| 15 | -1.607542 | 1.028983 | 2.668932 | 0.028725 | -4.632684 | 1.650265 | 3.827132 | 4.125967 | 1.419820 | 1.114608 |
| 16 | -1.479387 | -0.720721 | -0.483241 | 0.666806 | -1.827622 | 1.145017 | 2.805525 | 2.863499 | 4.868076 | 1.345775 |
| 17 | -1.989917 | -1.739324 | -0.738740 | 0.048710 | -0.727861 | 2.467566 | 1.668225 | 4.026885 | 3.470131 | 1.604032 |
| 18 | -1.905660 | -1.333194 | -0.874867 | 0.178728 | -0.813207 | 1.064777 | 2.820059 | 4.921509 | 7.533596 | 1.758663 |
| 19 | 0.393000 | -0.747532 | 2.781662 | 0.306974 | -4.688072 | 0.595645 | 1.914505 | 3.311281 | 7.665209 | 1.388369 |
Let's double-check to make sure our solutions fall within the problem variable boundaries.
population.min() > D_lower
0 True 1 True 2 True 3 True 4 True 5 True 6 True 7 True 8 True 9 True dtype: bool
population.max() < D_upper
0 True 1 True 2 True 3 True 4 True 5 True 6 True 7 True 8 True 9 True dtype: bool
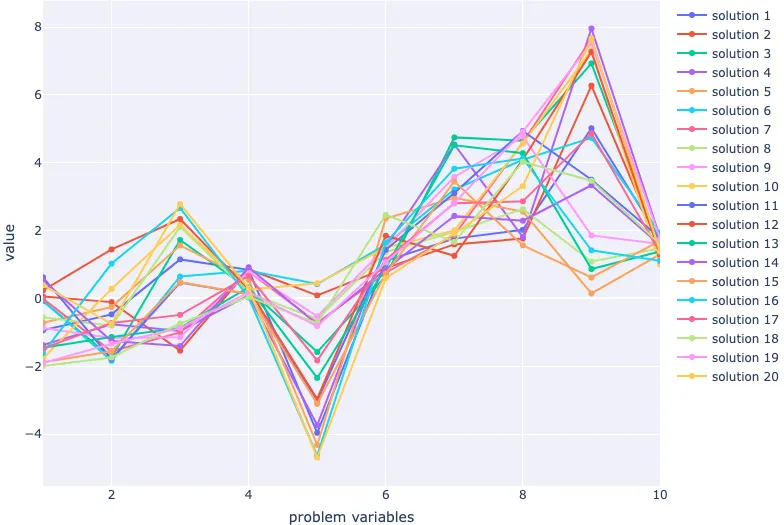
Great! Now all that's left is to visualise our population in the decision space. We'll use a parallel coordinate plot.
fig = go.Figure(
layout=dict(
xaxis=dict(title="problem variables", range=[1, 10]),
yaxis=dict(title="value"),
)
)
for index, row in population.iterrows():
fig.add_scatter(
x=population.columns.values + 1, y=row, name=f"solution {index+1}"
)
fig.show()
To compare one variable to another, we may also want to use a scatterplot matrix.
fig = px.scatter_matrix(population, title=" ")
fig.update_traces(diagonal_visible=False)
fig.show()
Conclusion
In this section, we had a closer look at multi-objective problems so that we knew how we could complete the initialisation stage in an evolutionary algorithm. We generated a population of solutions within upper and lower boundaries, checked to make sure the problem variables fell between the boundaries, and then visualised them using a scatterplot matrix and parallel coordinate plot. In a simple evolutionary algorithm, we have a population that is ready to enter the generational loop.