We are taking a practical approach in the following sections. As such, we need the right tools and environments available in order to keep up with the examples and exercises. We will be using Python 3 along with typical packages from its scientific stack, such as numpy (for multi-dimensional containers), pandas (for DataFrames), plotly (for plotting), etc. These instructions are written for Tensorflow v1, so you will see some specific package versions listed throughout for compatibility. We will write all of our code within a Jupyter Notebook, but you are free to use other IDEs such as PyCharm or Spyder.
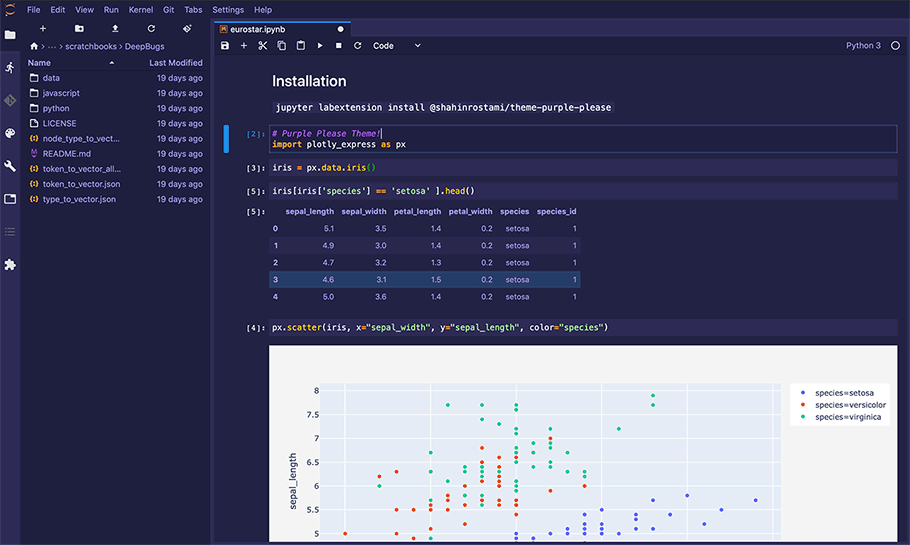
Figure 1 - A Jupyter Notebook being edited within Jupyter Lab.
Theme from https://github.com/shahinrostami/theme-purple-please
Get Anaconda
There are many different ways to get up and running with an environment that will facilitate our work. One approach I can recommend is to install and use Anaconda.
Anaconda® is a package manager, an environment manager, a Python/R data science distribution, and a collection of over 1,500+ open source packages. Anaconda is free and easy to install, and it offers free community support.
— https://docs.anaconda.com/anaconda/
To get up and running, just visit the Anaconda website and download a version of Anaconda for your operating system. I recommend getting the latest Anaconda version (2019.07 at the time of writing this section), and selecting a Python 3.X version.
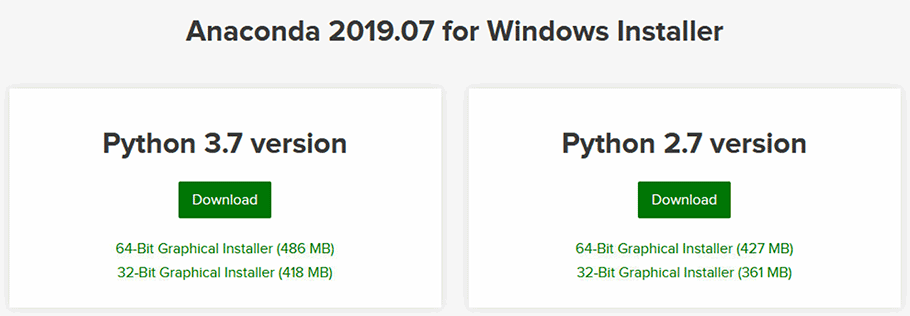
Figure 2 - Downloading an Anaconda Distribution for your operating system.
Create Your Environment
Once Anaconda is installed, we need to create and configure our environment. Again, there are many ways to accomplish this. You can complete all the steps using the Anaconda Navigator (graphical interface), but we will use the command-line interface, simply because it will give us a better report if and when something goes wrong.
If you added Anaconda to your PATH environment during the installation process, then you can run these commands directly from Terminal, Powershell, or CMD. If you didn't, then you can search for and run Anaconda Prompt.
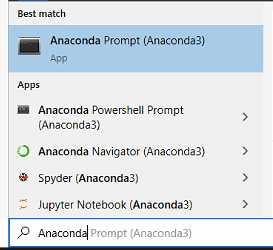
Figure 3 - Searching for Anaconda Prompt on Windows 10.
Now we can create and configure our conda environment using the following commands.
conda create -n ml python=3.6
This will create a conda environment named ml with the latest Python 3.6 package ready to go. You should be presented with a list of packages that will be installed, and asked if you wish to proceed. To do so, just enter the character y. If this operation is successful, you should see the following output at the end:
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
#
# To activate this environment, use
#
# $ conda activate ml
#
# To deactivate an active environment, use
#
# $ conda deactivate
As the message suggests, you will need to type the following command to activate and start entering commands within our environment named ml.
conda activate ml
Once you do that, you should see your terminal prompt now leads with the environment name within parantheses:
(ml) melica:~ shahin$
Note
The example above shows the macOS machine name "melica" and the user "shahin". You will see something different on your own machine, and it may appear in a different format on a different operating system such as Windows. As long as the prompt leads with "(ml)", you are on the right track.
This will allow you to identify which environment you are currently operating in. If you restart your machine, you should be able to use conda activate ml within your Anaconda prompt to get back into the same environment.
Install Packages
If you environment was already configured and ready, you would be able to enter the command jupyter lab to launch an instance of the Jupyter Lab IDE in the current directory. However, if we try that in our newly created environment, we will receive an error:
(ml) melica:~ shahin$ jupyter lab
-bash: jupyter: command not found
So let's fix that. Let's install a few packages we know we will be needed:
- Jupyter Lab
- Numpy
- Pandas
- Plotly
- Tensorflow
We will do them one-by-one to get a better idea of any errors if they occur. This time we will include the -y option which automatically says "yes" to any questions asking during the installation process.
conda install jupyterlab -y
conda install numpy=1.16 -y
conda install pandas -y
conda install plotly -y
You could do this in a single command, e.g.:
conda install jupyterlab numpy=1.16 pandas plotly -y
Let's install nodejs. This is needed to run our Jupyter Lab extension in the next section.
conda install -c conda-forge nodejs -y
Finally, let's install Tensorflow v1.
conda install -c conda-forge tensorflow=1
Install Jupyer Lab Extensions
There's one last thing we need to do before we move on, and that's installing any Jupyter Lab extensions that we may need. One particular extension that we need is the plotly extension, which will allow our Jupyter Notebooks to render our Plotly visualisations. Within your conda environment, simply run the following command:
jupyter labextension install @jupyterlab/plotly-extension
This may take some time, especially when it builds your jupyterlab assets, so keep an eye on it until you're returned control over the anaconda prompt, i.e. when you see the following:
(ml) melica:~ shahin$
Now we're good to go!
A Quick Test
Let's test if everything is working as it should be. In your anaconda prompt, within your conda environment, run the following command
jupyter lab
This should start the Jupyter Lab server and launch a browser window with the IDE ready to use.
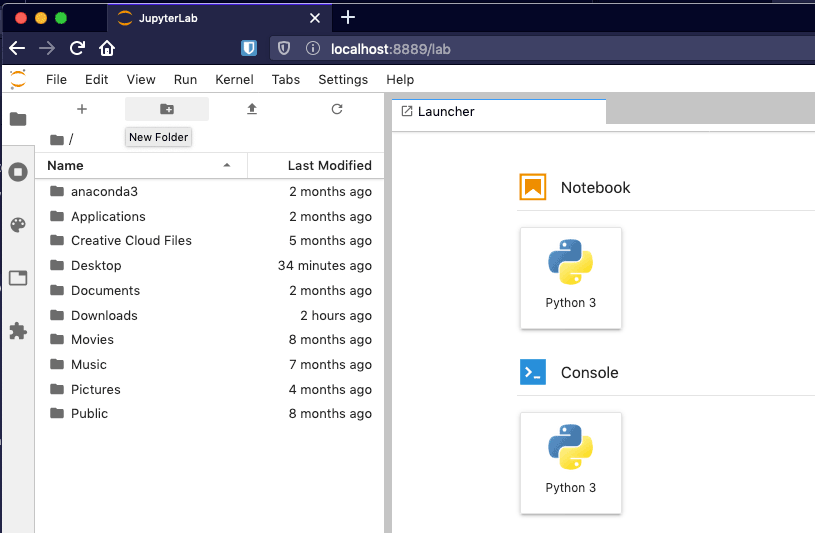
Figure 4 - A fresh installation of Jupyter Lab.
Let's create a new notebook. In the Launcher tab which has opened by default, click "Python 3" under the Notebook heading. This will create a new and empty notebook named Untitled.ipynb in the current directory.
Let's try to import our packages. If everything is configured as it should be, you should see no errors. Type the following into the first cell and click the "play" button to execute it and create a new cell.
import numpy as np # for multi-dimensional containers
import pandas as pd # for DataFrames
import plotly.express as px # plotly express
from tensorflow import keras # neural networks API
If we followed all the instructions and didn't encounter any errors, everything should be working. The last two lines beginning with pio.templates are modifications made to the colours/margins of all figures rendered by Plotly, and do not need to be included.
Enter the following code into the new and empty cell (below the first one) to test if Plotly visualisations will render within the notebook.
tips = px.data.tips()
fig = px.histogram(tips, x="total_bill", color="sex")
fig.show()
If the Jupyter Lab extension is installed and functioning, you should see an interactive histogram.
Summary
In this section we've downloaded, installed, configured, and tested our environment such that we're ready to run the following examples and experiments. If you ever find that you're missing packages, you can install them in the same way as we installed numpy and the others in this section.
