Preamble
import numpy as np # for multi-dimensional containers
import pandas as pd # for DataFrames
import plotly.graph_objects as go # for data visualisation
import plotly.express as px
Introduction
In this section, we're going to visualise Novel Coronavirus 2019 time series data for confirmed cases, recovered cases and deaths. We'll be working on two visualisations:
- A static map visualising our features for the latest time group in the dataset.
- An interactive and animated map visualising our features over time.
We'll be using the Mapbox service so we'll need to set our access token.
px.set_mapbox_access_token(access_token)
Note
To plot on Mapbox maps with Plotly you will need a Mapbox account and a public Mapbox Access Token. Copy yours over the string assigned to access_token in the cell above.

The Dataset
We're going to be using the Novel Corona virus - COVID19 dataset with the following description:
The new strain of Coronavirus has had a worldwide effect. It has affected people from different countries. The dataset provides, a time series data tracking the number of people affected by the virus, how many deaths has the virus caused and the number of reported people who have recovered.
Let's download it from their repository and take a peak.
data_url = pd.read_csv(
"https://datacrayon.com/datasets/time-series-19-covid-combined.csv"
)
data.head()
| Date | Country/Region | Province/State | Lat | Long | Confirmed | Recovered | Deaths | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2020-01-22 | Afghanistan | NaN | 33.0 | 65.0 | 0 | 0.0 | 0 |
| 1 | 2020-01-23 | Afghanistan | NaN | 33.0 | 65.0 | 0 | 0.0 | 0 |
| 2 | 2020-01-24 | Afghanistan | NaN | 33.0 | 65.0 | 0 | 0.0 | 0 |
| 3 | 2020-01-25 | Afghanistan | NaN | 33.0 | 65.0 | 0 | 0.0 | 0 |
| 4 | 2020-01-26 | Afghanistan | NaN | 33.0 | 65.0 | 0 | 0.0 | 0 |
We can see that we have the following features to work with.
data.columns.values
array(['Date', 'Country/Region', 'Province/State', 'Lat', 'Long',
'Confirmed', 'Recovered', 'Deaths', 'Active'], dtype=object)-
Province/State and Country/Region. These features contain the named location information associated with the sample. We'll use the Province/State field in our on-hover tool-tip and to group our colour grouping. We can see from the first five samples that some of these may be empty, or
NaN, so as a workaround we'll copy in the "Country/Region" feature where the data is missing.
missing_states = pd.isnull(data["Province/State"])
data.loc[missing_states, "Province/State"] = data.loc[
missing_states, "Country/Region"
]
- Lat and Long. These features contain the latitude and longitude geographic coordinates associated with the sample. We'll use both of these to determine where on the map we will draw our markers.
- Date. This feature contains the date associated with the sample. We'll use this to build our animation over time.
- Confirmed, Recovered, Deaths. These features contain numerical values for the number of confirmed cases, the number of recovered cases, and the number of deaths, respectively. We'll use the number of confirmed cases to change the size of our markers, and the number of deaths to change the colour.
We'll also add our own feature to estimate the number of active cases. We'll calculate it by subtracting the number of recovered cases and deaths from the confirmed cases. We can use this instead of the confirmed cases for our marker size in the animation.
data["Active"] = data["Confirmed"] - data["Recovered"] - data["Deaths"]
There's a possibility we will have NaN values in our data. We're not interested in investigating this further or conducting any imputation in this section, we will simply remove any rows that have this issue.
data = data.dropna()
The Latest Information
Let's create the first of our two visualisations, this one will present our features for the most recent time point in our data. We need to create a Boolean mask so that we can select only the relevant samples.
date_mask = data["Date"] == data["Date"].max()
date_mask
0 False
1 False
2 False
3 False
4 False
...
15161 False
15162 False
15163 False
15164 False
15165 True
Name: Date, Length: 14074, dtype: boolWe can now use this mask to select a subset of our dataset to produce a Figure object with Plotly Express.
fig = px.scatter_mapbox(
data[date_mask],
lat="Lat",
lon="Long",
size="Confirmed",
size_max=50,
color="Deaths",
color_continuous_scale=px.colors.sequential.Pinkyl,
hover_name="Province/State",
mapbox_style="dark",
zoom=1,
)
Reading through the parameters, we can see that we've:
* Passed in the masked subset of our dataset;
* Specified the latitude and longitude columns of that DataFrame to position our markers
* Set the size of our markers to be the number of confirmed cases, with a maximum size of 50.
* Set the colour of our markers to the number of deaths, on a continuous scale with the colour palette "Pinkyl";
* Set our on-hover tooltip to be the Province/State value;
* and set our map style to dark with a zoom of 1.
One extra configuration change we'll make is to remove the axis scale that will appear to the right of the figure. This is just a case of preference, you can see what you think of it by removing the line below or setting the value to True instead.
fig.layout.coloraxis.showscale = False
All that's left is to display our visualisation.
fig.show()
You can interact with the visualisation by dragging, zooming, hovering, etc.
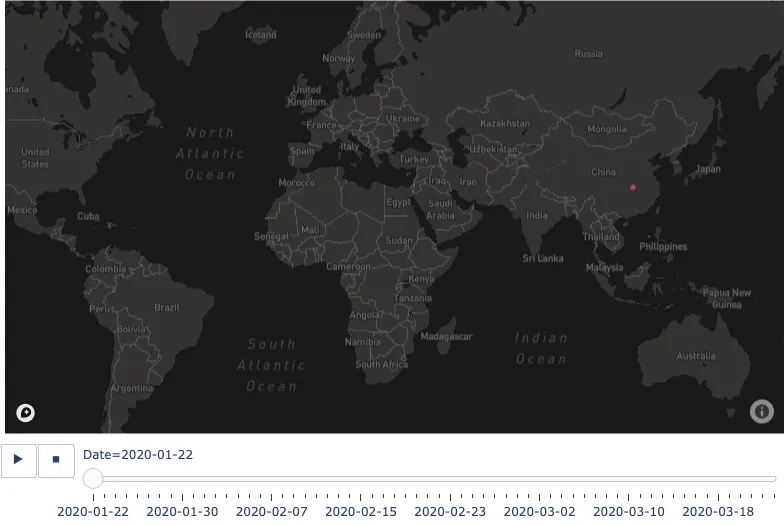
The Animated Time Series
Let's create the second of our two visualisations, this one will take us on a journey through time, giving us some idea of how the features have changed throughout the duration of the dataset. This time we will be passing in the entire DataFrame instead of the masked one, and we'll use our own active cases feature instead of the number of confirmed cases.
fig = px.scatter_mapbox(
data,
lat="Lat",
lon="Long",
size="Active",
size_max=50,
color="Deaths",
color_continuous_scale=px.colors.sequential.Pinkyl,
hover_name="Province/State",
mapbox_style="dark",
zoom=1,
animation_frame="Date",
animation_group="Province/State",
)
We can see two additional parameters for this plot which are used to specify how the animation frames are generated, and how they are grouped from frame to frame. In addition to removing the axis scale, we'll also make some additional changes to customise our animation and the positioning of some of the control elements.
fig.layout.updatemenus[0].buttons[0].args[1]["frame"]["duration"] = 200
fig.layout.updatemenus[0].buttons[0].args[1]["transition"]["duration"] = 200
fig.layout.coloraxis.showscale = False
fig.layout.sliders[0].pad.t = 10
fig.layout.updatemenus[0].pad.t = 10
Now we can display our visualisation.
fig.show()
You will be able to navigate the visualisation using the slider or click the play button to watch the animation. If you keep your attention focussed around China, you will notice the number of active cases grow and shrink over time.
Conclusion
In this section, we used a dataset capturing some features of the COVID19 cases to create static and animated map visualisations. To achieve the output, we used a few tools and services, primarily Plotly and Mapbox. The second visualisation, in particular, is quite interesting to watch as the number of active cases grow and shrink over time.