Preamble
from plotapi import Chord
Introduction
Note
This document is best viewed online where the interactivity of the demonstrations can be experienced.
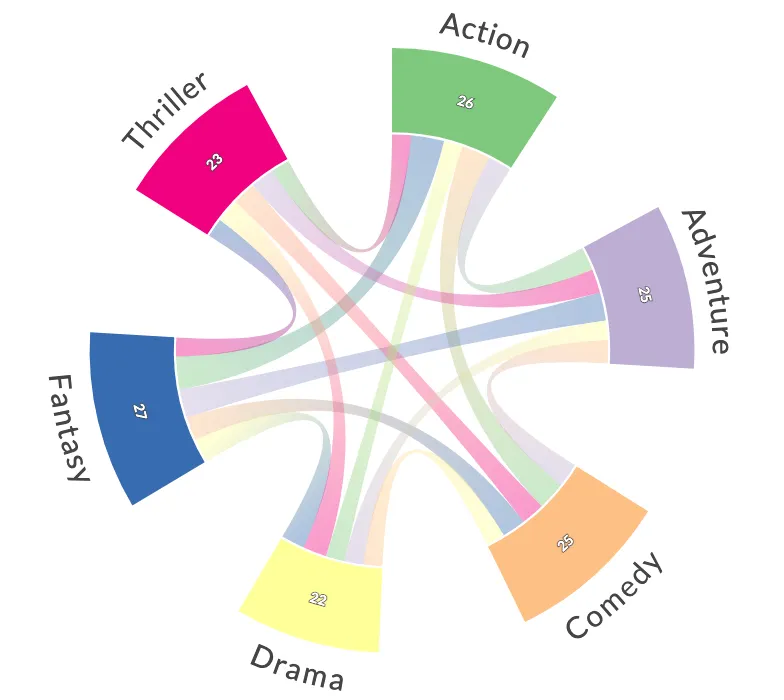
In a chord diagram (or radial network), entities are arranged radially as segments with their relationships visualised by arcs that connect them. The size of the segments illustrates the numerical proportions, whilst the size of the arc illustrates the significance of the relationships1. Chord diagrams are useful when trying to convey relationships between different entities, and they can be beautiful and eye-catching.
Get PlotAPI for Chord Diagrams
Click here to get access to the full-featured chord visualization API, producing beautiful interactive visualizations, e.g. those featured on the front page of Reddit.
License
To use the full-featured plotapi package, you need to assign a valid license key. This can be purchased here.
Chord.api_key("your-api-key")
We'll use the following data for the co-occurrence matrix and names parameters until we cover divided diagrams.
matrix = [
[0, 5, 6, 4, 7, 4],
[5, 0, 5, 4, 6, 5],
[6, 5, 0, 4, 5, 5],
[4, 4, 4, 0, 5, 5],
[7, 6, 5, 5, 0, 4],
[4, 5, 5, 5, 4, 0],
]
names = ["Action", "Adventure", "Comedy", "Drama", "Fantasy", "Thriller"]
Defaults
Without passing in any arguments for the customisation parameters, the output will use the default value.
Chord(matrix, names)
Outputs Methods
Chord Pro supports the following outputs.
HTML to Jupyer Lab Cell (Interactive)
Outputs the interactive diagram to a Jupyter Lab cell.
Chord(matrix, names)
HTML to file (Interactive)
Saves an interactive HTML file locally.
Chord(matrix, names).to_html("out.html")
PNG to Jupyer Lab Cell (Image)
Outputs a PNG to a Jupyter Lab Cell.
Chord(matrix, names).show_png()
PNG to file (image)
Saves a PNG file locally.
Chord(matrix, names).to_png("out.png")
PDF to file
Saves a PDF file locally.
Chord(matrix, names).to_pdf("out.pdf")
Disable SSL Verification
Chord.verify_ssl(True)
Some users behind school/corporate networks may experience issues contacting the Chord API end-points. One workaround is to use Chord.verify_ssl(False). You will receive an InsecureRequestWarning with a link to further information.
Chord.verify_ssl(False)
Chord(matrix, names, title="SSL Verification Disabled")
Chord Colours
colors="rainbow"
You can select a color scheme from the following: ['league', 'monsters', 'movies', 'category10', 'accent', 'dark2', 'paired', 'pastel1', 'pastel2', 'set1', 'set2', 'set3', 'tableau10', 'rainbow', 'sinebow', 'yellow_red', 'yellow_brown', 'yellow_green', 'yellow_blue', 'red_purple', 'purple_red', 'purple_blue', 'orange_red', 'green_blue', 'blue_purple', 'blue_green', 'cubehelix', 'cool', 'warm', 'cividis', 'plasma', 'magma', 'inferno', 'viridis', 'turbo', 'brown_green', 'purple_green', 'pink_green', 'red_blue', 'red_grey', 'red_yellow_blue', 'red_yellow_green', 'spectral', 'blues', 'greens', 'greys', 'oranges', 'purples', 'reds']
Chord(matrix, names, colors="league")
The colors parameter also accepts a Python list of HEX colour codes.
grayscale = [
"#222222",
"#333333",
"#4c4c4c",
"#666666",
"#848484",
"#9a9a9a",
]
Chord(matrix, names, colors=grayscale)
Opacity
opacity=0.8
This sets the opacity for the arcs when they are not selected (mouseover/touch).
Chord(matrix, names, opacity=0.2)
Reverse Gradients
opacity=0.8
This sets the direction of the arc gradients.
Chord(matrix, names, reverse_gradients=True)
Arc Numbers
arc_numbers=False
This sets the visibility of quantity labels on segments.
Chord(matrix, names, arc_numbers=True)
Diagram Title
titles=""
This sets the text and visibility of the diagram title.
Chord(matrix, names, title="Movie Genre Co-occurrence")
Padding
padding=0.01
This sets the padding between segments as a fraction of the circle.
Chord(matrix, names, padding=0.5)
Width
width=700
This sets the width (and height) of the chord diagram.
Chord(matrix, names, width=400)
Rotation
rotate=0
This sets the rotation of the chord diagram.
Chord(matrix, names, rotate=-30)
Radius Scales
inner_radius_scale=0.45,
outer_radius_scale=1.1,
This sets the inner and outer radius scale of the chord diagram.
Chord(matrix, names, inner_radius_scale=0.3, outer_radius_scale=1.5)
Label Color
label_colors="#454545"
This sets the label colour. It can be a string or a list of strings.
Chord(matrix, names, label_colors="#B362FF")
Curved Labels
curved_labels=False
This turns curved labels on or off. It's not suitable for diagrams with many segments, but sometimes it can improve the look.
Chord(matrix, names, curved_labels=True)
Label Wrapping
wrap_labels=True
This turns label wrapping on or off. It's on by default.
Let's temporarily change our names data to demonstrate this.
names = [
"Exciting Action",
"Fun Adventure",
"Hilarious Comedy",
"Drama",
"Fantasy",
"Chilling Thriller",
]
Chord(matrix, names)
Now to demonstrate label wrapping set to disabled.
Chord(matrix, names, wrap_labels=False)
We'll restore our names data before we continue.
names = ["Action", "Adventure", "Comedy", "Drama", "Fantasy", "Thriller"]
Margins
margin=100
This sets the chord diagram margin.
Chord(matrix, names, margin=200)
Font-size
font_size="16px"
font_size_large="20px"
This sets the font-size for two views, font-size-large (@media min-width: 600px), otherwise font-size. Setting a large font-size may require adjustment of the margin parameter to avoid text-clipping.
Chord(matrix, names, font_size="20px", font_size_large="30px")
Popup Width
popup_width=350
This sets the max-width of the popup.
Chord(matrix, names, popup_width=150)
Popup Text
conjunction="and"
verb="occur together in"
noun="instances"
These change parts of the popup text:

e.g. we could change it to:

Chord(
matrix,
names,
conjunction="&",
verb="appear together in",
noun="cases",
)
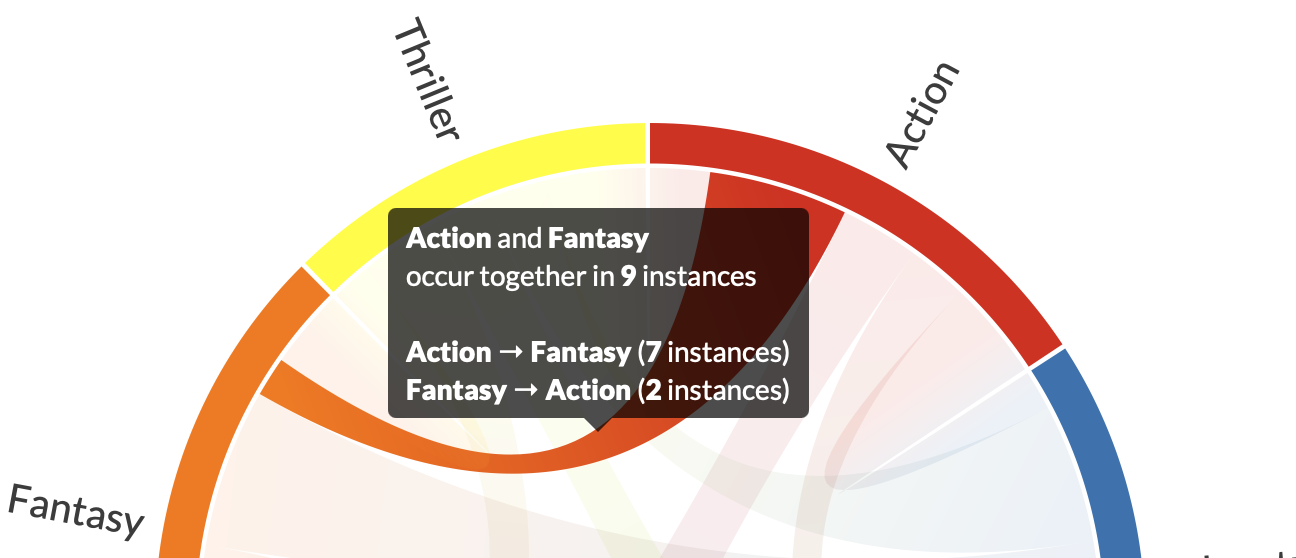
Symmetric and Asymmetric Diagrams
symmetric=True
This turns the symmetric mode on or off. The primary support is for symmetric diagrams, but there is some support for asymmetric ones.
For example, this will update the popup text to reflect the asymmetry of relationships.
matrix = [
[0, 5, 3, 4, 7, 3],
[2, 0, 6, 5, 2, 7],
[6, 3, 0, 4, 5, 7],
[8, 3, 4, 0, 4, 7],
[2, 8, 6, 7, 0, 4],
[5, 3, 3, 4, 2, 0],
]
Chord(matrix, names, symmetric=False)
Download Button
allow_download=False
This sets the visibility of the Download button. This uses client-side scripts to create and download an SVG from the current visualisation. This has been tested on Firefox and Safari.
Chord(matrix, names, allow_download=True)
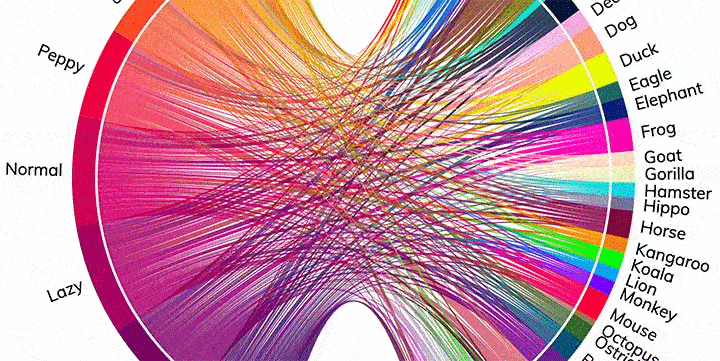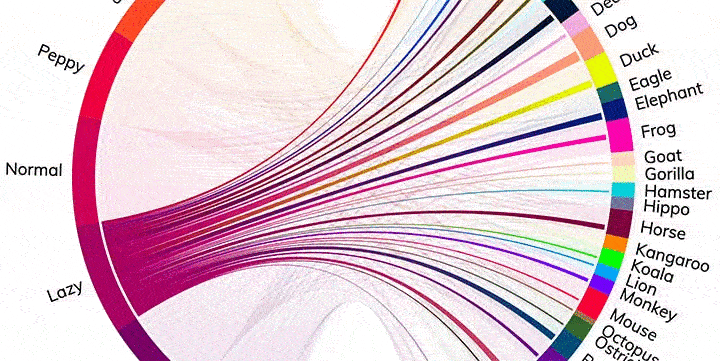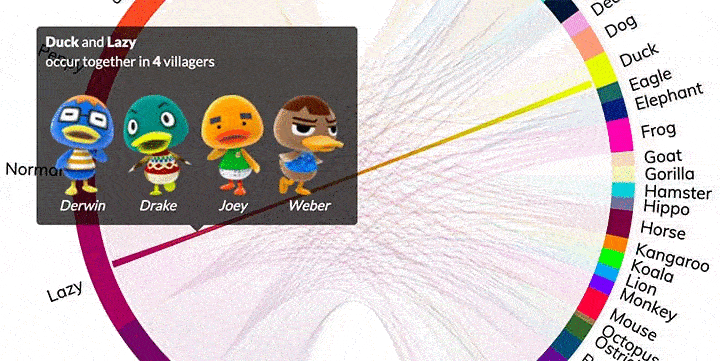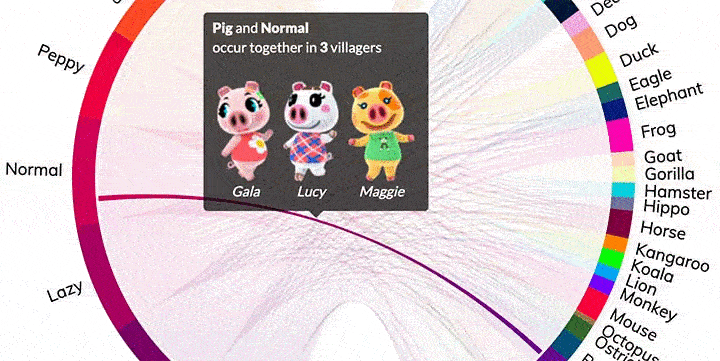
Popup Details, Thumbnails, and Custom HTML
details=[],
details_thumbs=[],
In the basic version of chord, matrix and names are the only sets of data that can be used to create a chord diagram. In the Pro version, you can also use details and details_thumbs. These enable the rich hover boxes.

First, we'll reduce the co-occurrence frequency in our matrix to make the following examples easier to follow.
matrix = [
[0, 2, 3, 1, 4, 1],
[2, 0, 2, 1, 3, 2],
[3, 2, 0, 1, 2, 2],
[1, 1, 1, 0, 2, 2],
[4, 3, 2, 2, 0, 1],
[1, 2, 2, 2, 1, 0],
]
Let's create and populate an example details and details_thumbs variable.
details = [
[[], ["Movie 1","Movie 2"], ["Movie 3","Movie 4","Movie 5"], ["Movie 6","Movie 7"], ["Movie 8","Movie 9","Movie 10","Movie 11"], ["Movie 12"]],
[["Movie 13","Movie 14"], [], ["Movie 15","Movie 16"], ["Movie 17"], ["Movie 18","Movie 19","Movie 20"], ["Movie 21","Movie 22"]],
[["Movie 23","Movie 24","Movie 25"], ["Movie 26","Movie 27"], [], ["Movie 28"], ["Movie 29","Movie 30"], ["Movie 31","Movie 32"]],
[["Movie 33"], ["Movie 34"], ["Movie 35"], [], ["Movie 36","Movie 37"], ["Movie 38","Movie 39"]],
[["Movie 40","Movie 41","Movie 42","Movie 43"], ["Movie 44","Movie 45","Movie 46"], ["Movie 47","Movie 48"], ["Movie 49","Movie 50"], [], ["Movie 51"]],
[["Movie 52"], ["Movie 53","Movie 54"], ["Movie 55","Movie 56"], ["Movie 57","Movie 58"], ["Movie 59"], []],
]
The details parameter can accept HTML.
details_thumbs = [
[[], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png"]],
[["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], [], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"]],
[["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], [], ["https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"]],
[["https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png"], [], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"]],
[["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], [], ["https://datacrayon.com/datasets/pp_img/3.png"]],
[["https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png","https://datacrayon.com/datasets/pp_img/3.png"], ["https://datacrayon.com/datasets/pp_img/3.png"], []],
]
It looks a bit messy, so let's achieve some clarity using the pandas.DataFrame HTML table output.
import pandas as pd
pd.DataFrame(details)
| 0 | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| 0 | [] | [Movie 1, Movie 2] | [Movie 3, Movie 4, Movie 5] | [Movie 6, Movie 7] | [Movie 8, Movie 9, Movie 10, Movie 11] | [Movie 12] |
| 1 | [Movie 13, Movie 14] | [] | [Movie 15, Movie 16] | [Movie 17] | [Movie 18, Movie 19, Movie 20] | [Movie 21, Movie 22] |
| 2 | [Movie 23, Movie 24, Movie 25] | [Movie 26, Movie 27] | [] | [Movie 28] | [Movie 29, Movie 30] | [Movie 31, Movie 32] |
| 3 | [Movie 33] | [Movie 34] | [Movie 35] | [] | [Movie 36, Movie 37] | [Movie 38, Movie 39] |
| 4 | [Movie 40, Movie 41, Movie 42, Movie 43] | [Movie 44, Movie 45, Movie 46] | [Movie 47, Movie 48] | [Movie 49, Movie 50] | [] | [Movie 51] |
| 5 | [Movie 52] | [Movie 53, Movie 54] | [Movie 55, Movie 56] | [Movie 57, Movie 58] | [Movie 59] | [] |
pd.DataFrame(details_thumbs)
| 0 | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| 0 | [] | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png] |
| 1 | [https://datacrayon.com/datasets/pp_img/3.png,... | [] | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png] | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... |
| 2 | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [] | [https://datacrayon.com/datasets/pp_img/3.png] | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... |
| 3 | [https://datacrayon.com/datasets/pp_img/3.png] | [https://datacrayon.com/datasets/pp_img/3.png] | [https://datacrayon.com/datasets/pp_img/3.png] | [] | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... |
| 4 | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [] | [https://datacrayon.com/datasets/pp_img/3.png] |
| 5 | [https://datacrayon.com/datasets/pp_img/3.png] | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png,... | [https://datacrayon.com/datasets/pp_img/3.png] | [] |
Chord(matrix, names, details=details, details_thumbs=details_thumbs)
Popup Thumbnails Spacing and Size
thumbs_width=85
thumbs_margin=5
thumbs_font_size=14
These allow changing the spacing and size of the details thumbnails.
Chord(
matrix,
names,
details=details,
details_thumbs=details_thumbs,
thumbs_width=40,
thumbs_margin=10,
thumbs_font_size=10,
)
Bipartite Chord Diagram
bipartite=False,
bipartite_idx=0,
bipartite_size=0.5,
-
bipartiteenables/disables the divided chord diagram mode. -
bipartite_idxsets the dividing point of the matrix. -
bipartite_sizesets the size of the spacing that creates the divide.
First, we'll change the data in matrix and names such that they're suitable for our bipartite chord diagram. We'll add some new colours too.
matrix = [
[0, 0, 0, 1, 4, 1],
[0, 0, 0, 1, 3, 2],
[0, 0, 0, 1, 2, 2],
[1, 1, 1, 0, 0, 0],
[4, 3, 2, 0, 0, 0],
[1, 2, 2, 0, 0, 0],
]
names = ["A", "B", "C", "1", "2", "3"]
colors = ["#7400B8", "#5E60CE", "#5684D6", "#56CFE1", "#64DFDF", "#80FFDB"]
Chord(matrix, names, colors=colors, bipartite=True, bipartite_idx=3)
Bipartite Chord Diagram Labels
bipartite_left_label="",
bipartite_right_label=""
These parameters allow you to label either side of the divided chord diagram.
Chord(
matrix,
names,
colors=colors,
bipartite=True,
bipartite_idx=3,
bipartite_left_label="Left Side",
bipartite_right_label="Right Side",
)
Colored Diagonals
colored_diagonals=True
This sets the visibility of the occurrences (not co-occurrences), i.e. the representation of the diagonal of the matrix.
names = ["one", "two", "three", "four", "five", "six"]
matrix = [
[19, 5, 6, 4, 7, 4],
[5, 4, 5, 4, 6, 5],
[6, 5, 0, 4, 5, 5],
[4, 4, 4, 0, 5, 5],
[7, 6, 5, 5, 0, 4],
[4, 5, 5, 5, 4, 0],
]
Chord(matrix, names, title="Diagonals coloured")
Chord(matrix, names, title="Diagonals not coloured", colored_diagonals=False)
Equal-sized Sections
This makes use of the colored_diagonals feature above. The idea is to determine the max frequency in our plot, and set all the diagonals to that value. Let's assume that is 20 for this example.
names = ["one", "two", "three", "four", "five", "six"]
matrix = [
[20, 0, 0, 0, 0, 0],
[0, 20, 0, 0, 0, 0],
[0, 0, 20, 0, 0, 0],
[0, 0, 0, 20, 0, 0],
[0, 0, 0, 0, 20, 0],
[0, 0, 0, 0, 0, 20],
]
Chord(matrix, names, title="Diagonals not coloured", colored_diagonals=False)
We can see above that this creates a chord diagram with equal-sized segments. All we need to do now is introduce our relationships, and subtract them from the corresponding diagonal value.
matrix = [
[11, 4, 5, 0, 0, 0],
[4, 14, 2, 0, 0, 0],
[5, 2, 13, 0, 0, 0],
[0, 0, 0, 20, 0, 0],
[0, 0, 0, 0, 20, 0],
[0, 0, 0, 0, 0, 20],
]
Chord(matrix, names, title="Diagonals not coloured", colored_diagonals=False)
We could add some padding to separate them too.
matrix = [
[11, 4, 5, 0, 0, 0],
[4, 14, 2, 0, 0, 0],
[5, 2, 13, 0, 0, 0],
[0, 0, 0, 20, 0, 0],
[0, 0, 0, 0, 20, 0],
[0, 0, 0, 0, 0, 20],
]
Chord(
matrix,
names,
title="Diagonals not coloured",
colored_diagonals=False,
padding=0.2,
)
Mix and Match
matrix = [
[0, 5, 6, 4, 7, 4],
[5, 0, 5, 4, 6, 5],
[6, 5, 0, 4, 5, 5],
[4, 4, 4, 0, 5, 5],
[7, 6, 5, 5, 0, 4],
[4, 5, 5, 5, 4, 0],
]
names = ["Action", "Adventure", "Comedy", "Drama", "Fantasy", "Thriller"]
Chord(
matrix,
names,
padding=0.5,
reverse_gradients=True,
colors="accent",
curved_labels=True,
label_color="#777777",
font_size_large=30,
inner_radius_scale=0.45,
outer_radius_scale=1.4,
arc_numbers=True,
margin=50
)
-
Tintarev, N., Rostami, S., & Smyth, B. (2018, April). Knowing the unknown: visualising consumption blind-spots in recommender systems. In Proceedings of the 33rd Annual ACM Symposium on Applied Computing (pp. 1396-1399). ↩